Fall 2017: Building Livability
UTA researchers are creating a more sustainable, affordable North Texas for the future.
Skip to content. Skip to main navigation.
UTA researchers are creating a more sustainable, affordable North Texas for the future.
From carbon dioxide conversion to landfill mining, researchers at UTA are seeking viable alternative energy options.
Found in everything from space shuttles to dental fillings, composite materials have thoroughly infiltrated modern society. But their potential is still greatly untapped, offering researchers ample opportunity for discovery.
Within the particle showers created at the Large Hadron Collider, answers to some of the universe’s mysteries are waiting.
Model systems like pigeons can help illuminate our own evolutionary and genomic history.
UT Arlington's tiny windmills are bringing renewable energy to a whole new scale.
The stability of our highways, pipelines, and even manholes is reaching a breaking point.
Scientists believe they have discovered a subatomic particle that is crucial to understanding the universe.
UT Arlington researchers unlock clues to the human body’s most mysterious and complex organ.
UT Arlington researchers probe the hidden world of microbes in search of renewable energy sources.
Wounded soldiers are benefiting from Robert Gatchel’s program that combines physical rehabilitation with treatment for post-traumatic stress disorder.
Tiny sensors implanted in the body show promise in combating acid reflux disease, pain and other health problems.
Nanotechnology researchers pursue hybrid silicon chips with life-saving potential.
Biomedical engineers combat diseases with procedures that are painless to patients.

Illustration by JEAN FRANCOIS PODEVIN
What does snake blood have to do with regenerating heart tissue in humans? How about water fleas with precision medicine? Frog eyes with evolution? And what do all these questions have in common?
They’re what a handful of researchers at The University of Texas at Arlington are trying to find out as they tackle important issues in the burgeoning, and increasingly critical, field of genomics. And with the opening of the North Texas Genome Center and its state-of-the-art equipment and ethos of collaboration and information-sharing, the answers to these and other potentially life-changing questions may soon be close at hand.
Genomics has opened a new realm of discovery for scientists, particularly those whose ultimate goal is to impact human health. Because researchers can't conduct most experiments on humans, they must use model systems, such as lab rats. But according to Todd Castoe, assistant professor of biology, there just aren't very many interesting phenotypes—or gene expressions—in some of these go-to model systems. However, advances in genome technology have changed the way researchers are able to conduct their experiments.
To that end, Dr. Castoe is studying the ability of some snakes to regenerate their organs, sometimes up to 100 percent, within two days of a large meal. During this period, the snakes make what would be toxic levels of insulin in humans, but remain unharmed. After about 10 days of digestion, the snakes then reverse that process, shrinking their organs to save energy.
Using genomic technology, Castoe sequenced the genome of the Burmese python and other snakes to see how close they are to the human genome. Through genome sequencing and comparative analysis, he knows the gene responsible for regulating insulin is comparable to the same gene in humans and can estimate how snake-like organ regeneration might be stimulated in humans. Without that comparison, he says, he would only be able to study the snake's ability, not whether it could be applicable to humans.
"Now that we know humans and snakes have similar sets of genes, the key question is why can't humans undergo regenerative growth—or can they?" Castoe says.
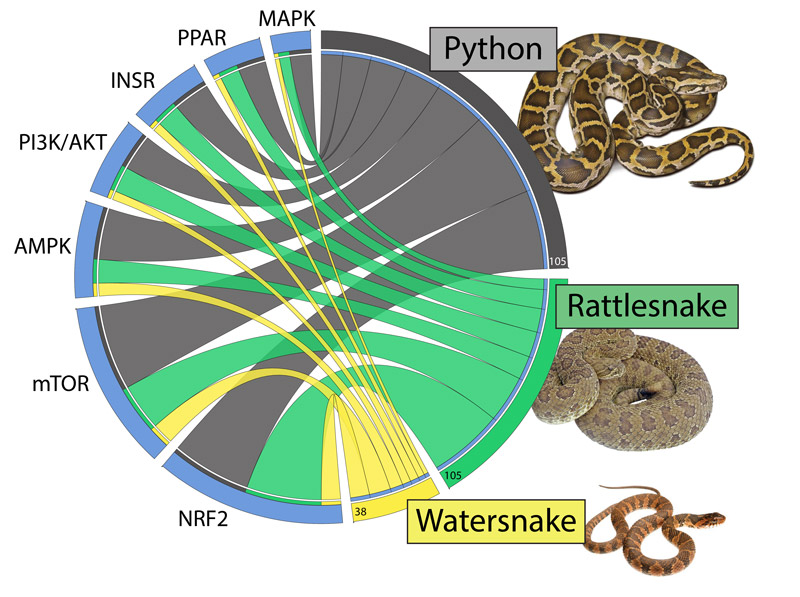
Todd Castoe's chart compares how gene expression across seven different pathways in snakes are used to control organ regeneration. Such information may be valuable for understanding human organ regeneration and cancer.
He is working to understand which snake genes and growth signals control rapid regeneration and then see whether humans can do the same thing. There are many potential health implications involved—whether humans could ever regrow heart tissue after a heart attack, for example, or whether we could use the snake's downregulation technique to shrink intestinal cancer.
To conduct these experiments, Castoe has partnered with researchers at the Massachusetts Institute of Technology to grow human organoids in petri dishes. After giving human stem cells growth signals, the cells develop into mini-organs. Then, using snake blood plasma on the organoids—which are as close to real human organs as he can create in the lab—Castoe takes samples of the organoid's RNA to understand what genes are being turned on or off and at what levels. He compares those responses to what happens in the snake and tries to understand the similarities and differences.
It's this application of genomic technology to new and valuable—albeit unusual—model systems that makes his research potentially useful in advancing human health. Thanks to genomics, Castoe and his colleagues have begun asking questions in a fundamentally different way when exploring new research interests.
"Don't limit yourself to only studying traditional model species. Don't rule out lots of intriguing biology to figure out what we can study," Castoe says. "Chase the biology you think is most important to understand. Don't even ask if you can do it—genomics allows you to do it."
Instead of snakes, biology Assistant Professor Matt Walsh is using Daphnia for his model system, which are small aquatic crustaceans commonly known as water fleas. He has found that these creatures can temporarily alter their gene expression depending on cues in their environment. The Daphnia convert these environmental signals into heritable changes not to the DNA sequence itself, but to chemical modifications of DNA in the genome—known as epigenetic modifications—that may alter patterns of gene expression for multiple generations.
Similar patterns of transgenerational plasticity have been observed in humans, so understanding these epigenetic changes has the potential to eventually benefit precision medicine, which uses molecular profiling to optimize treatments for individual patients.
"In humans, we have known for years that environmental and behavioral factors can have substantial effects on individuals, as well as their offspring for multiple generations, due to changes the environment can have on epigenetic modifications of the genome that alter gene expression," explains Castoe, who is collaborating with Dr. Walsh on the project. "But these factors are not currently incorporated into modern medicine, and approaches to understand the epigenome and its relevance to medicine represent a major frontier for up-and-coming genomics research."
In a related project, Walsh recently received a five-year, $600,000 grant from the National Science Foundation for his work to "resurrect" Daphnia eggs from the 1990s and observe their genetic modifications when they encountered stressors in their environment. He plans to take a sample sediment core to collect layers of viable, unhatched eggs on the bottom of northern U.S. lakes. Then, he will isotopically age each layer, going back in time to observe how Daphnia gene expression changed after the arrival of new invasive predators. It's a way to effectively observe phenotypic changes through time.
The project is a novel approach toward answering a longstanding question in evolutionary biology—whether the ability of an organism to modify gene expression and phenotype based on its environment accelerates or slows genetic adaptation.
"Scientists have argued about this for more than 100 years now," Walsh says. "The work done in my lab should have broad interest across the field of evolution."
Walsh's and Castoe's genomics work and that of researchers across the North Texas region will receive a big boost when UTA opens the North Texas Genome Center (NTGC) this year, bringing massive genomics computing capabilities to campus.
Jon Weidanz, associate vice president for research and the NTGC's interim director, says the center's new leading-edge analyzers will provide the most advanced DNA and RNA sequencing available. This capability provides the foundation for data-driven discovery, one of the themes of UTA's Strategic Plan 2020: Bold Solutions | Global Impact. The center will also advance research in human and animal studies, contributing to two other strategic themes: health and the human condition and global environmental impact.
"Technology is playing a greater role in health care and treating disease," Dr. Weidanz says. "With high throughput sequencing of a patient's tumor, we'll know what the specific targets are and we can make decisions on the best way to help that person. That's the heart of the matter: personalized medicine."
The NTGC will be available to any researchers who want to collaborate on projects and take advantage of the research capabilities at UTA.
Duane Dimos, vice president for research, says the NTGC reinforces UTA as a quickly growing research institution.
"This is a huge opportunity as biotech and health science research starts to grow in the DFW metroplex," he says. "Building this center not only supports our faculty, but also attracts academic and corporate partners."
For his genomics research, Corey Roelke is staying close to home. The biology lecturer just concluded two large grants from Texas Parks and Wildlife Department and the Comptroller of Public Accounts. He and Assistant Professor Matt Fujita studied the genetic composition of the spot-tailed earless lizard Holbrookia lacerata, which has experienced a steady decline in population in Central and South Texas. By analyzing sequences of the lizards' DNA, they found evidence that these populations are two distinct species, and the southern group isn't doing very well from a preservation standpoint.
"This matters because if they are two different species, they are treated independently of each other by the federal government," Dr. Roelke says. "One is far more imperiled, but many people would prefer them not to be on the endangered species list because it could possibly impact industrial development."
In addition to this project, Dr. Fujita is studying frog vision and what part environmental factors play in its evolution. Because vision plays a key role in how an organism interacts with its environment, he hypothesizes that nocturnal frogs and those that live underground won't see all the same colors as diurnal frogs living in other surroundings.
Fujita plans to sequence eye genes in about 100 different types of frogs. With the data, he'll be able to analyze the frogs' gene expression—whether they're using vision genes in the same way, depending on where they live.
"Our discoveries could provide important insight not just in frog vision, but also in how vision could evolve in other species," he says.
The project is funded by a $714,000 NSF grant as well as more than $300,000 from the Natural Environment Research Council in the United Kingdom. When the four-year grant wraps up in 2021, Fujita and his co-researchers will display the results at the Smithsonian National Museum of Natural History as well as the Natural History Museum in London.
UTA’s new center is set to become a hub for innovation in precision medicine, biotech, and more.
The ability to rapidly sequence genomes, DNA, and RNA is providing doctors, scientists, and engineers critical new tools in health care delivery and research, changing the way that doctors treat patients. UTA, in partnership with the University of North Texas Health Science Center (UNTHSC), is launching the North Texas Genome Center (NTGC) to provide massive genome sequencing capabilities to enhance health care delivery and support the rapidly expanding biotech sector in North Texas and the surrounding six-state region.
Set to be housed in UTA's new 229,000-square-foot Science & Engineering Innovation & Research building, the NTGC will feature five state-of-the-art, high-throughput genome sequencers from Illumina. As one of only a few centers in the central United States featuring NovaSeq6000s, the NTGC will be able to meet the rising demand for whole genome sequencing in the region, serving as a hub for advancing collaborative, cutting-edge medical research and lowering the cost for high-quality human whole genome sequencing. The service will be open for clinical customers, drug developers, and academic researchers in March 2018.
Excepting accidents, genomic factors play a role in nine of the 10 leading causes of death in the United States. Whole genome sequencing is becoming the foundation for precision health, a rapidly growing field that examines differences in people's genetic makeup to provide more personalized and effective health care solutions.
Campus investments in new faculty and new federal and state research grants are positioning UTA as a regional leader in developing the platform technologies needed for precision medicine to become a reality. UTA will partner with hospitals and medical systems throughout the region to support the needs of their clinicians for whole genome, whole exome, and other patient genomic data.
"The new center aligns with UTA's strategic focus on both health and the human condition and data-driven discovery and will consolidate existing and future programs in genomics, computational sciences, and genetic counseling," says President Vistasp Karbhari. "The NTGC will also catalyze the University's emergence as a leader in precision health and the transformation of the region into a high-tech science hub."
Jon Weidanz, associate vice president for research and interim director of the NTGC, emphasizes the importance of the partnership with UNTHSC on this project.
"It's a real differentiator, as it means that we can combine the engineering, science, and nursing expertise at UTA with the clinical health experience of UNTHSC to produce innovative health care solutions," he says.
UTA expects the NTGC to boost the state's economy through patented inventions, startups, and job creation.
"UTA is a hub for innovation in North Texas, and the North Texas Genome Center will increase our capacities in this area," says Duane Dimos, vice president for research and NTGC associate director.